Notes
Sort by:| Metadata | Start Date | End Date | Comment |
|---|---|---|---|
|
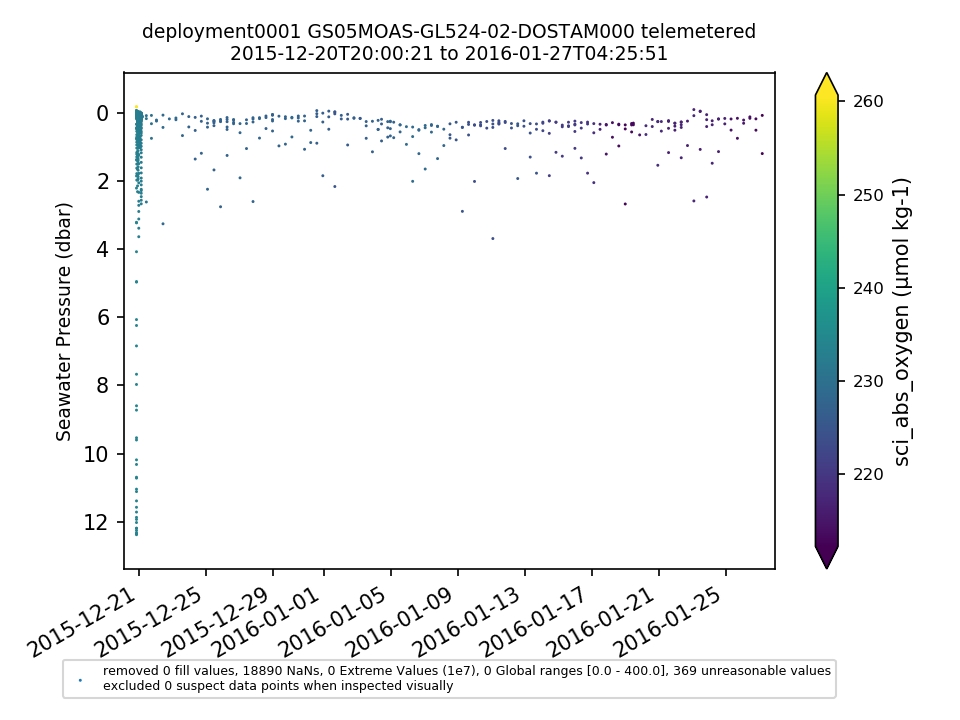
GS05MOAS-GL524-02-DOSTAM000 |
|||
|
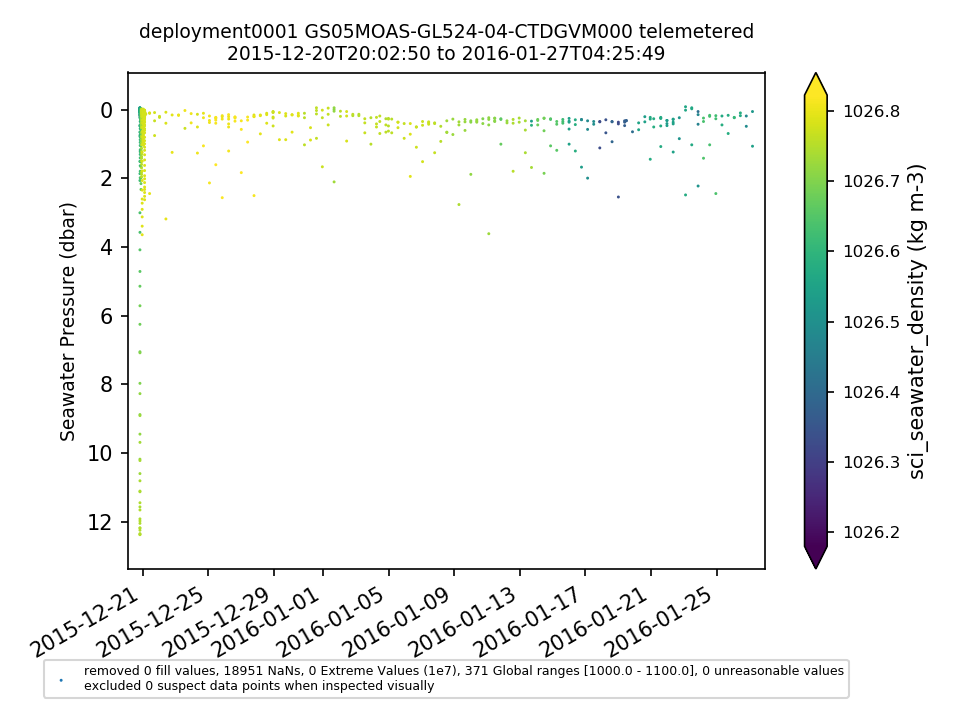
GS05MOAS-GL524-04-CTDGVM000 |
|||
|
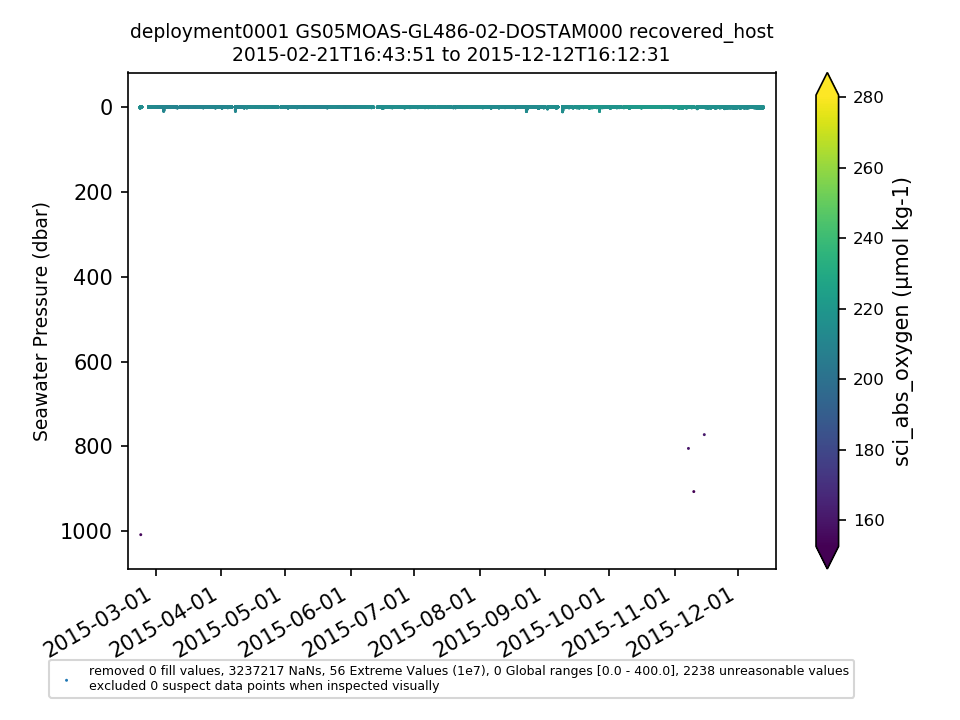
GS05MOAS-GL486-02-DOSTAM000 |
|||
|
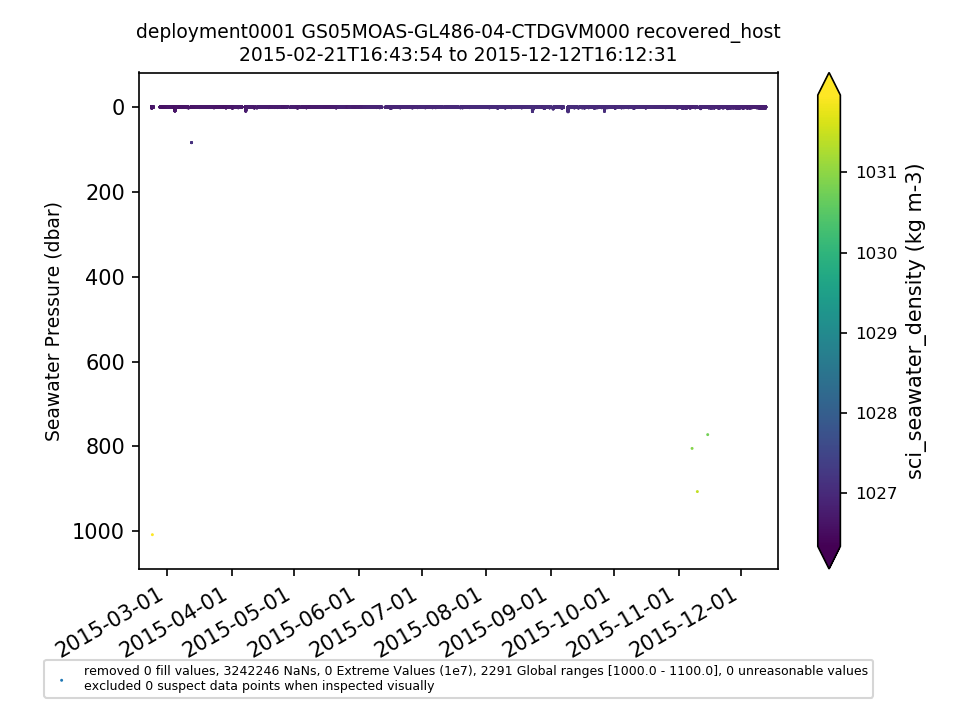
GS05MOAS-GL486-04-CTDGVM000 |
|||
|
GS05MOAS-GL485-02-DOSTAM000 |
92% of sci_abs_oxygen values are NaNs. Lat and lon are parameters used in the data product algorithm, so I think this got messed up with the re-organization of the glider lat/lon parameters. See Redmine ticket #14486 By Lori Garzio, on 1/23/20 |
||
|
GS05MOAS-GL485-04-CTDGVM000 |
97% of sci_seawater_density values are NaNs. Lat and lon are parameters used in the data product algorithm, so I think this got messed up with the re-organization of the glider lat/lon parameters. See Redmine ticket #14486 By Lori Garzio, on 1/23/20 |
||
|
GS05MOAS-GL485 Deployment: 1 |
recovered_host data aren't available for this deployment. According to the ingest csv, the glider failed and recovered data aren't expected. This dataset should be annotated. By Lori Garzio, on 1/22/20 |
||
|
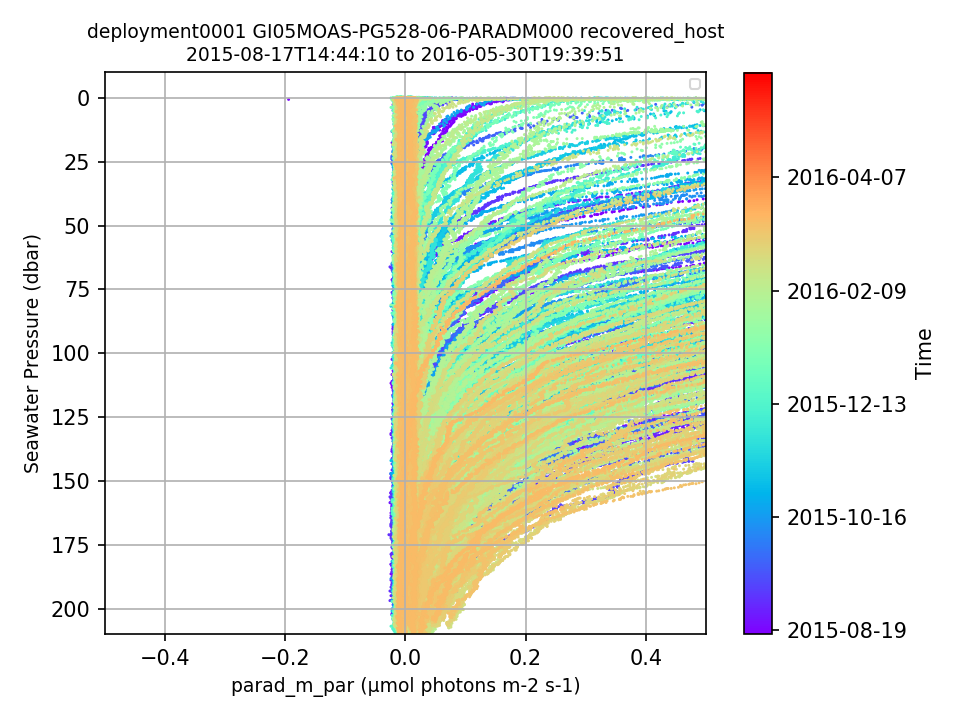
GI05MOAS-PG528-06-PARADM000 Deployment: 1 |
97% of the sci_bsipar_par values for this deployment are outside of global ranges. sci_bsipar_par is the unscaled L0 PAR data product. The global ranges for this parameter are the same as for the scaled L1 PAR data product [0.0, 2500.0]. The global range for sci_bsipar_par should be reviewed. By Lori Garzio, on 1/22/20 |
||
|
GI05MOAS-PG528-06-PARADM000 Deployment: 1 |
|||
|
GI05MOAS-PG528-04-FLORTO000 |
There are no global ranges in the system for sci_bb3slo_b470_units, flort_o_bback_b470_total, flort_o_bback_b532_total, sci_bb3slo_b660_units, and flort_o_bback_b660_total. By Lori Garzio, on 1/22/20 |
||
|
GI05MOAS-PG528-04-FLORTO000 |
The arrays for the following variables are all NaNs: sci_bb3slo_b470_units, flort_o_bback_b470_total, flort_o_bback_b532_total, sci_bb3slo_b660_units, flort_o_bback_b660_total. According to the provenance.json (attached), the following cal values are missing: CC_1_dark_counts_volume_scatter, CC_1_scale_factor_volume_scatter, CC_1_measurement_wavelength, CC_2_dark_counts_volume_scatter, CC_2_scale_factor_volume_scatter, CC_2_measurement_wavelength, CC_3_dark_counts_volume_scatter, CC_3_scale_factor_volume_scatter, and CC_3_measurement_wavelength. By Lori Garzio, on 1/22/20 |
||
|
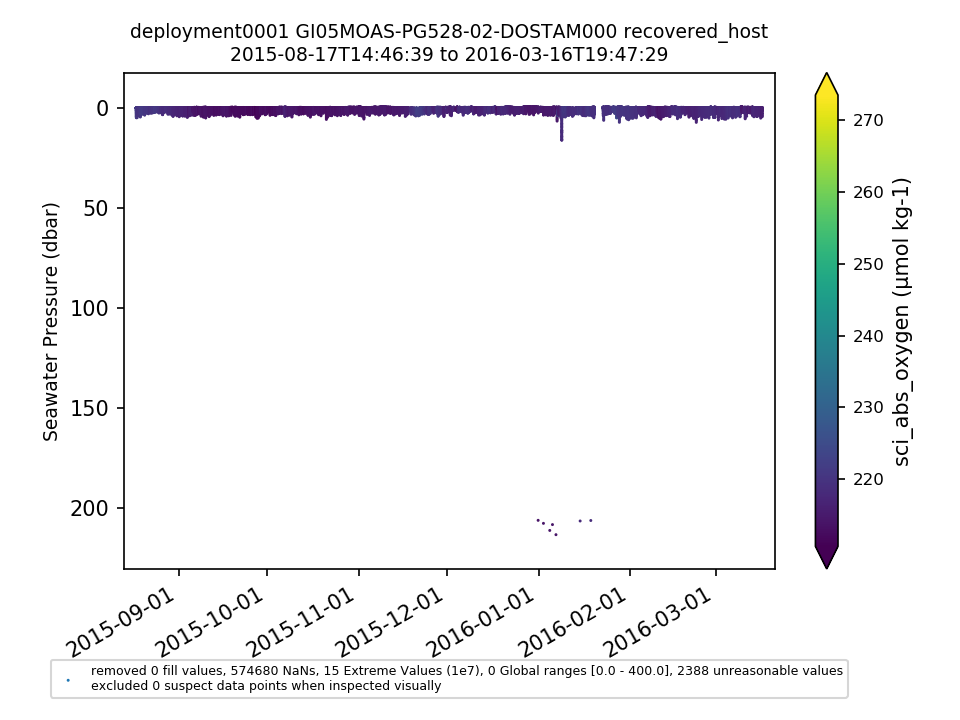
GI05MOAS-PG528-02-DOSTAM000 |
|||
|
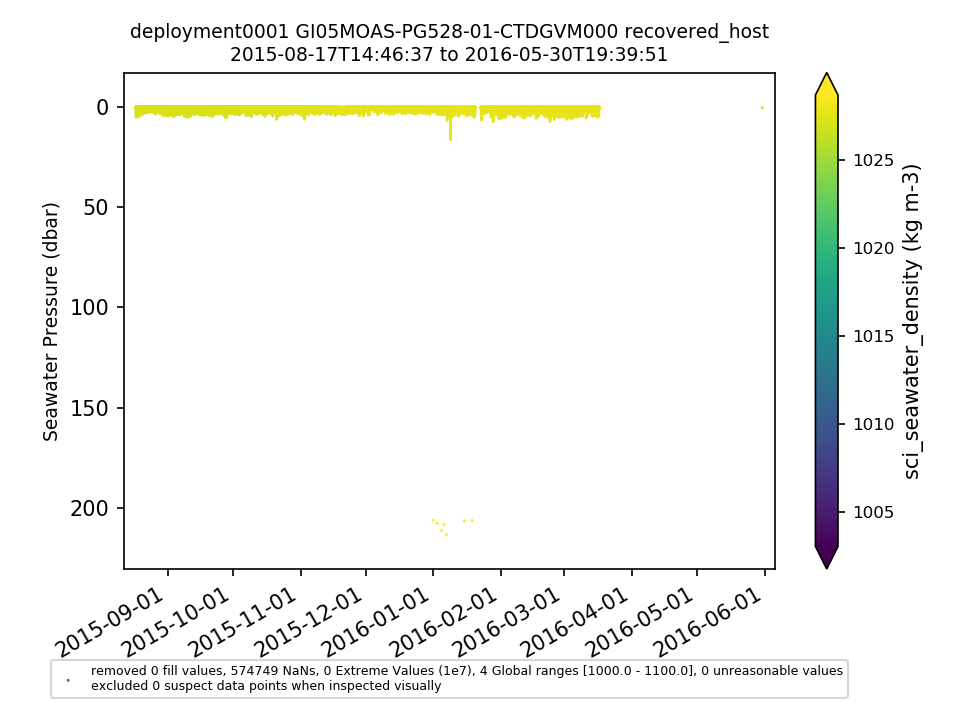
GI05MOAS-PG528-01-CTDGVM000 |
|||
|
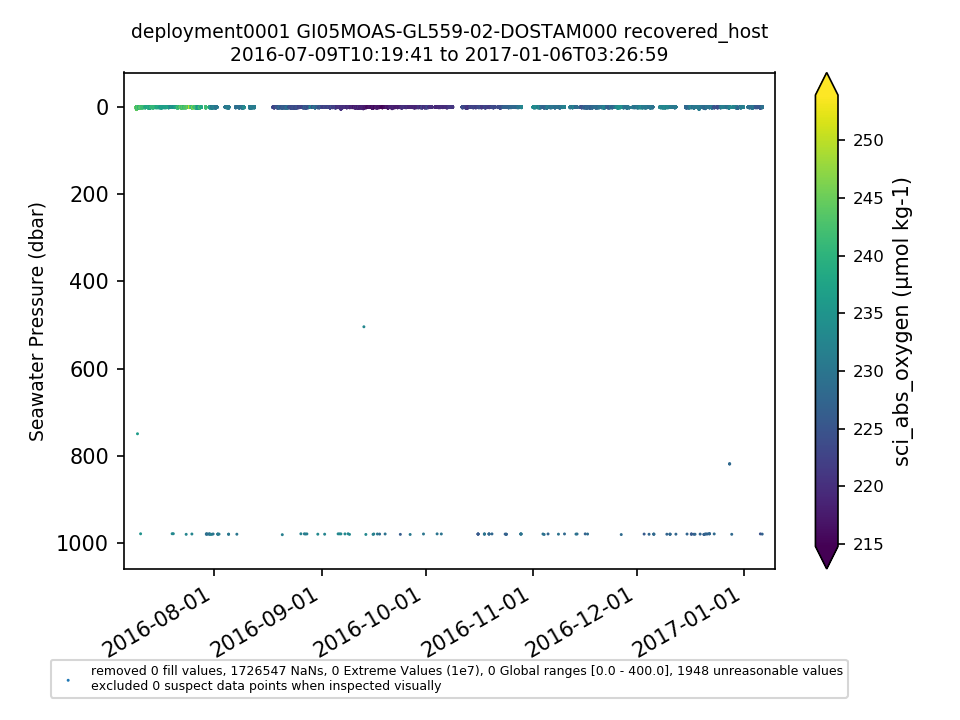
GI05MOAS-GL559-02-DOSTAM000 |
|||
|
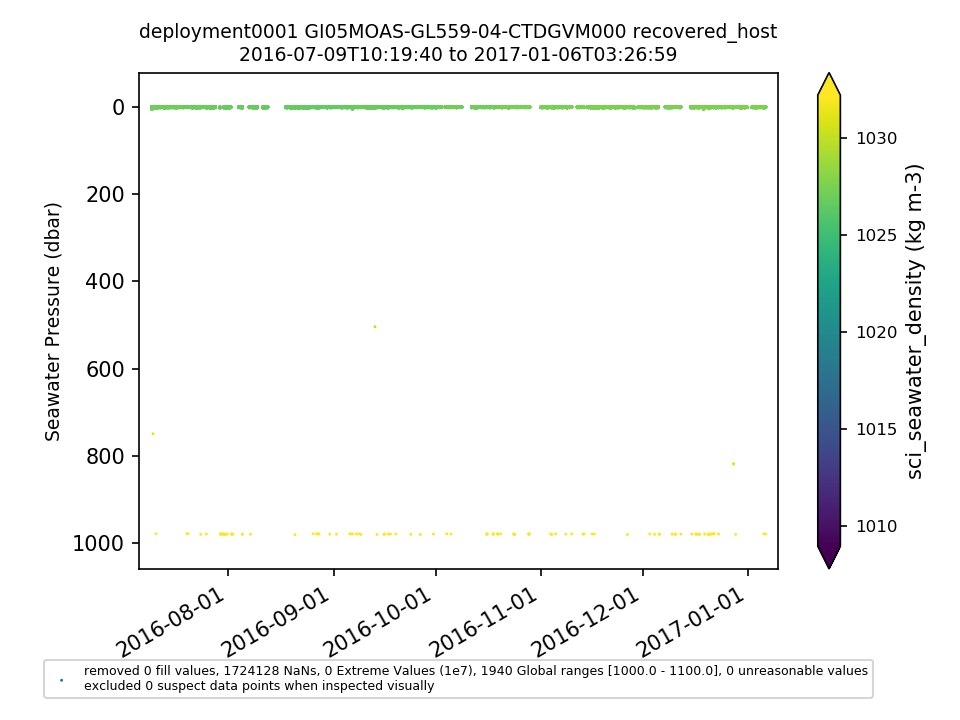
GI05MOAS-GL559-04-CTDGVM000 |
|||
|
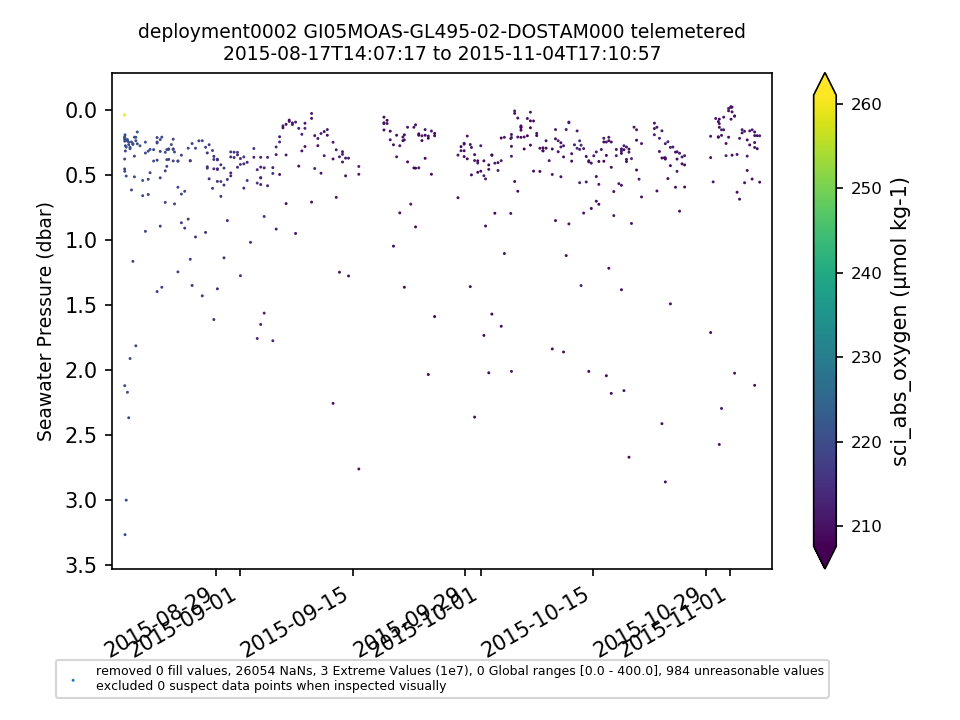
GI05MOAS-GL495-02-DOSTAM000 |
|||
|
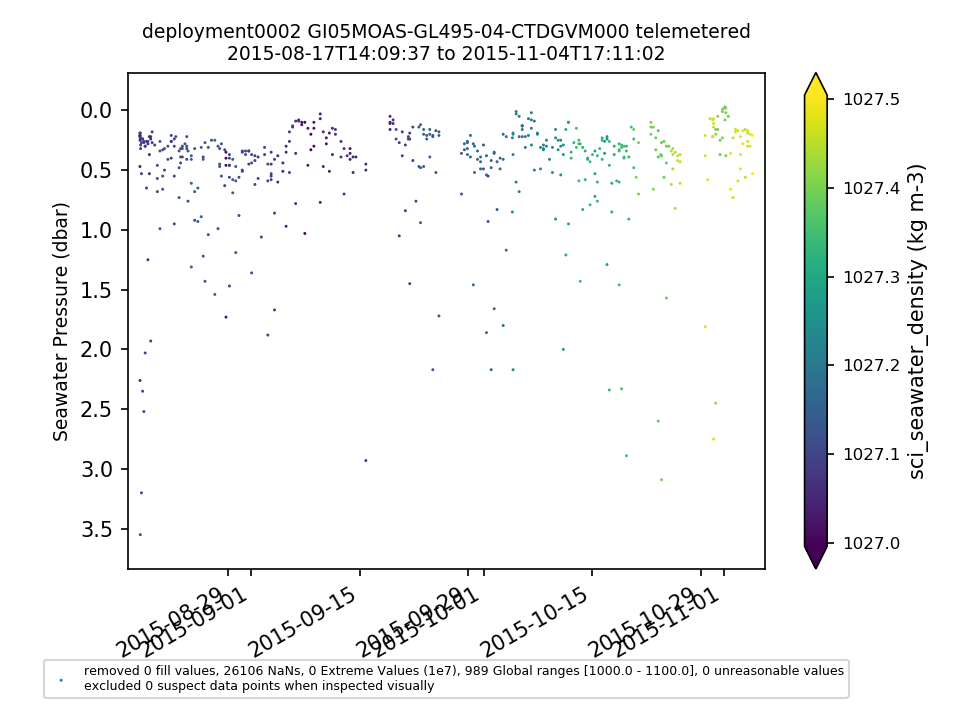
GI05MOAS-GL495-04-CTDGVM000 |
|||
|
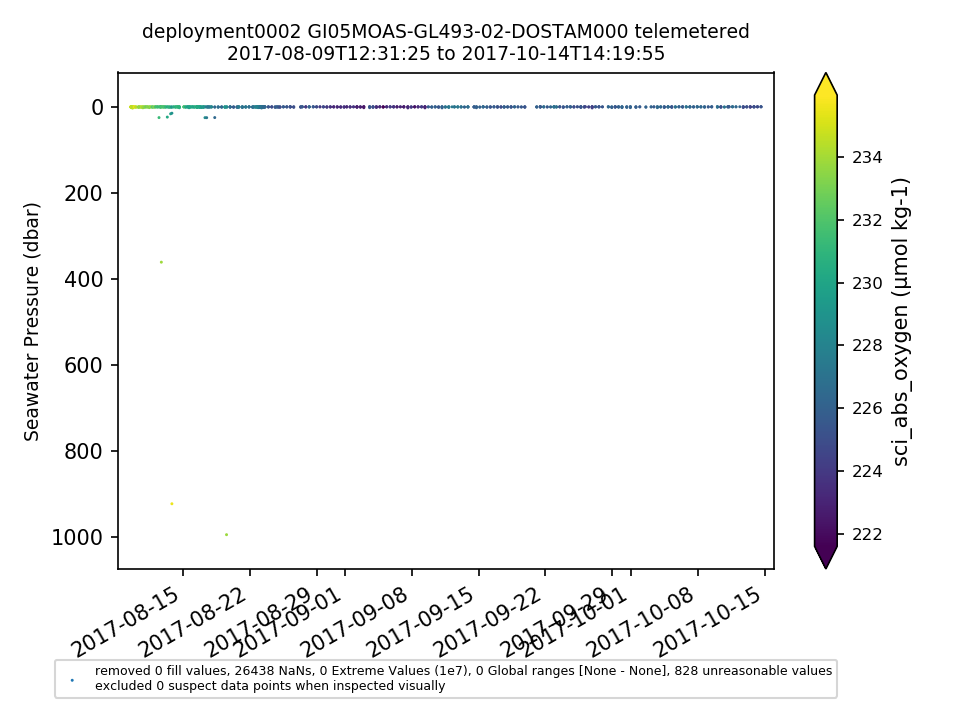
GI05MOAS-GL493-02-DOSTAM000 |
|||
|
GI05MOAS-GL493-04-CTDGVM000 |
|||
|
GI05MOAS-GL493 Deployment: 2 |
There is no end date in the system for this glider (as of 1/13/2020). It was deployed >2 years ago on 8/9/2017. Also, recovered data aren't available for download. The recovered data need to be ingested or the dataset should be annotated to explain why recovered data aren't available. By Lori Garzio, on 9/23/19 |